
The file is divided into sections where each section corresponds to the data. NMRbox is a resource for biomolecular NMR (Nuclear Magnetic Resonance) software.

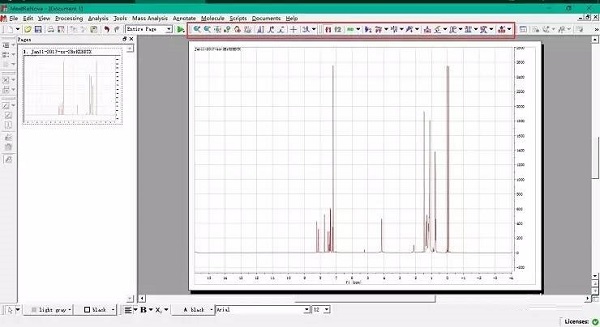
It provides tools for finding the software you need, documentation and tutorials for getting the most out of the software, and cloud-based virtual machines for executing the softwareĬombined automated NOE assignment and structure determination module (CANDID) is a new software for efficient NMR structure determination of proteins by automated assignment of the NOESY spectra. First released in 2008, UNIO is the ultimate result of more than 15 years of meticulous research performed in order to guarantee accurate, objective and highly automated protein structure determination by NMRĬANDID uses an iterative approach with multiple cycles of NOE cross-peak assignment and protein structur The UNIO multi-purpose software suite enables you to conduct unsupervised NMR data analysis for biomolecular 3D structure determination and more. Prices were checked November 2013, are for academic licenses, and are in US dollars unless stated otherwise. Work up noesy in mestrenova trial#ĢD structure drawing: PERCH NMR Software Tools: Windows: 3 month trial version: focused on small molecules: Avogadro: Windows, MacOSX, Linux: protein structural analysis with residual dipolar couplings: TALOS-N. The essential steps of an NOE-based structure determination by NMR are data acquisition, signal identification, chemical shift assignment, NOE assignment, and structure calculation. 4 Current software like UNIO 6, 7 or FLYA 8, 9 performs the computational steps in an almost fully automated manner Structure Determination Using NMR.


 0 kommentar(er)
0 kommentar(er)
